|
|
|
|

|
|
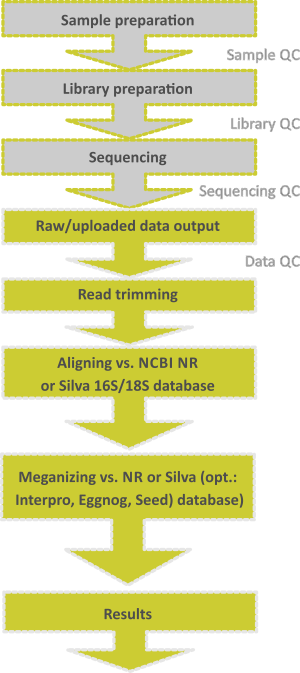
The term metagenomics has been defined as “The study of DNA from uncultured organisms” (Jo Handelsman). A genome is the entire genetic information of one organism, whereas a metagenome is the entire genetic information of an ensemble of organisms. It is a culture-independent tool for studying bacterial, viral, fungal or eukaryotic communities and can be divided in two main categories.
- One is to focus on specific marker genes for taxonomic identification only. For this so-called targeted amplicon approach, the ribosomal small subunit RNA (16S / 18S rRNA) gene has emerged as the most used marker.
- Alternatively, the use of the entire DNA or RNA for sequencing is often referred to as whole genome shotgun (WGS) metagenomics offering a random representation of all extracted genomic sequences, giving deep insights into the
taxonomic and metabolic profiles of these communities. In addition to the information about taxonomic diversity (who is there), shotgun metagenomics gives insight into the physiology of the organisms present in the environment (what are they doing). It has been estimated that >99% of the microbial numbers in nature are non-culturable by available techniques. Hence, new cultivation-independent methods to study the function and diversity of microorganisms in nature are needed. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale and detail than before.
Applications
-
Rapid viral- and microbial diagnosing, tropical disease screening
-
Human microbiomics
-
Water and soil testing
-
Marine metagenomics
-
Animal and plant disease diagnostics
-
Food safety
-
Petroleum-based microbiology
-
Biofuel
-
Sterility monitoring
-
Detection of contamination
Packages
Targeted amplicon 16S/18S metagenomics, only taxonomical profiling
→ Package 1: 1–48 samples
→ Package 2: 49–96 samples
→ Package 3: 97–192 samples
Whole genome shotgun metagenomics, taxonomical & functional profiling
→ Package 1: 1–20 samples
→ Package 2: 21–50 samples
→ Package 3: 50 < samples
Optional: Metagenomics de novo assembly, taxonomical & functional profiling
→ Package 1: 1–20 samples
→ Package 2: 21–50
→ Package 3: 50
Optional: Viral genome finishing and annotation, SNV analysis
Seqomics’ professional metagenomics services include:
→ Free of charge project consultation, study design
→ Discussion about the appropriate technical approach and platform for the project
→ Detailed consultation on sampling options, coverage estimation
→ Metagenome isolation (total DNA or RNA), optional
→ Professional sample processing throughout the workflow
→ Expert bioinformatics service for data analysis
→ Comprehensive, publication-ready reporting of data and results
→ Continuous communication with customers, follow-up the project
Links
- MEGAN6 for Windows
- MEGAN6 for MacOS
- MEGAN6 for Linux
- MEGAN6 manual
- MEGAN6 Community
- Illumina Miseq platform
- iontorrent system
 |




 WinSCP (Windows)
WinSCP (Windows) Commander One (Mac OS)
Commander One (Mac OS)